Pancreatic cancer is the third leading cause of cancer-related deaths in the U.S., with 80 to 85% of cases diagnosed too late for effective treatment. The disease’s silent progression and anatomical complexity make it difficult for human radiologists to detect early on—which led Johns Hopkins researchers to turn to a new solution: artificial intelligence.
To train AI models capable of detecting this cancer early enough to make a difference in patients’ survival, JHU researchers including Malone Center member Zongwei Zhou and Bloomberg Distinguished Professor Alan Yuille worked with NVIDIA researchers and institutions around the world to launch the Pancreatic Tumor Segmentation Dataset, or PanTS—the largest and most comprehensive CT scan dataset ever released for pancreatic cancer detection.
They presented the fully open-source dataset at the 39th Annual Conference on Neural Information Processing Systems, held December 2–7 in San Diego.
PANTs was built using MONAI Label, NVIDIA’s open-source AI framework for medical imaging. Radiologists used MONAI to perform interactive 3D segmentation, enabling scalable, human-in-the-loop annotation workflows. All told, the dataset contains over 36,000 3D CT scans from 145 medical centers, with expert-validated annotations of over 993,000 anatomical structures, including the pancreas, tumors, and surrounding organs.
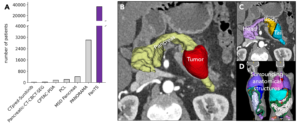
Dataset characteristics and visualization.
Aside from this impressive size and diversity, each CT scan in the dataset includes rich metadata—such as patient age, sex, diagnosis, and more—and information about imaging protocols and biomarkers to help enable AI models capable of identifying high-risk individuals across populations and imaging conditions, the researchers say.
“Although current AI isn’t yet ready for cancer screening in a general population, if we use imaging biomarkers, clinical notes, and deep neural networks to select high-risk patients, we can transform a blunt population-level screener into a precision cancer detection tool,” explains Zhou, the senior author of the project.
Another improvement PanTS boasts is its inclusion of a large number of CT scans without pancreatic tumors. According to the research team, it’s important to include these scans to reduce the prevalence of false positives flagged by oversensitive AI models, which can lead to unnecessary patient anxiety, overdiagnosis, and costly follow-ups.
With these enhancements, PanTS drastically improves AI accuracy in pancreatic tumor detection. According to the researchers, models trained on PanTS significantly outperform those trained on existing public datasets. These gains are directly attributable to PanTS’s scale and anatomical richness, they say.
And because PanTS has been made public, with a reserved test set for third-party validation, AI developers and hospitals around the world can train models to spot pancreatic cancer earlier.
“Our team will keep promoting open science in medical computer vision—especially for cancer research, where public annotated datasets are painfully limited,” says Wenxuan Li, a PhD student of computer science and the first author on the project. “We hope PanTS can serve as an exemplar open dataset in the medical domain, following the impact of BraTS, LiTS, and KiTS, and we’re excited to continue incorporating community suggestions to make PanTS even better.”
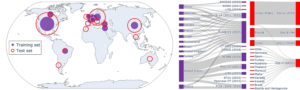
Geographic diversity of the PanTS dataset.
With contributions from institutions across Europe, Asia, and North America, PanTS represents a global effort to accelerate AI innovation in oncology, radiotherapy planning, and surgical decision support—but the team welcomes additional testing from institutions on underrepresented continents such as Africa, South America, and Australia to improve the generalizability of the dataset.
By enabling earlier and more accurate tumor detection, PanTS has the potential to improve survival rates and transform pancreatic cancer care, the researchers say. In fact, a new algorithm currently being developed by Johns Hopkins researchers is reportedly detecting pancreatic cancer in CT scans over a year earlier than most human radiologists would be able to—all because of the training data provided by PanTS.
“Looking forward, we want to detect all types of cancers at earlier stages, classify them, and predict how they will develop over time,” says Zhou. “Ultimately, we envision a world where we can predict, well in advance, who is at high risk for cancer, enabling personalized screening and timely early detection far before the cancer has a chance to spread.”
Additional authors from Johns Hopkins include graduate students Xinze Zhou and Qi Chen; Tianyu Lin, Engr ’25 (MS); incoming postdoctoral fellow Pedro R.A.S. Bassi; and School of Medicine faculty Kai Ding and Heng Li. Core contributors from NVIDIA include Yucheng Tang and Daguang Xu.
