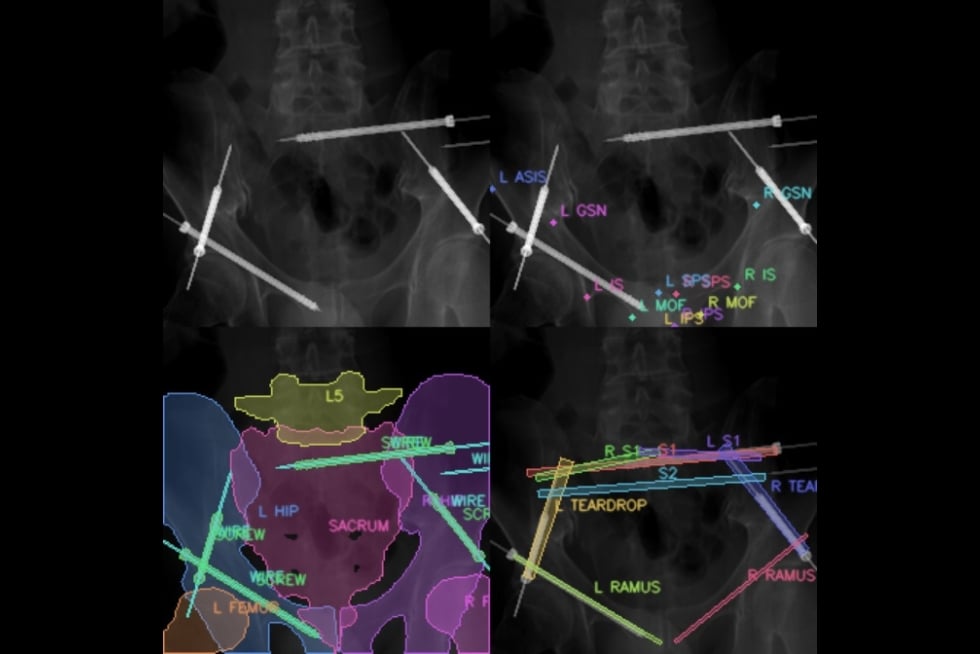
Researchers at the Johns Hopkins University are leveraging the power of machine learning to improve X-ray-guided pelvic fracture surgery, an operation to treat an injury commonly sustained during car crashes.
A team of researchers from the university’s Whiting School of Engineering and the School of Medicine plan to increase the efficiency of this surgery by applying the benefits of surgical phase recognition, or SPR, a cutting-edge machine learning application that involves identifying the different steps in a surgical procedure to extract valuable insights into workflow efficiency, the proficiency of surgical teams, error rates, and more. The team presented its X-ray-based SPR-driven approach, called Pelphix, last month at the 26th International Conference on Medical Image Computing and Computer-Assisted Intervention in Vancouver.
“Our approach paves the way for surgical assistance systems that will allow surgeons to reduce radiation exposure and shorten procedure length for optimized pelvic fracture surgeries,” said research team member Benjamin Killeen, a doctoral candidate in the Department of Computer Science and a member of the Advanced Robotics and Computationally AugmenteD Environments (ARCADE) Lab.
SPR lays the foundation for automated surgical assistance and skill analysis systems that promise to maximize operating room efficiency. While SPR typically analyzes full-color endoscopic videos taken during surgery, it has to date ignored X-ray imaging—the only imaging available for many procedures, such as orthopedic surgery, interventional radiology, and angiology, leaving these procedures unable to reap the benefits of SPR-enabled advancements.
Despite the rise of modern machine learning algorithms, X-ray images are still not routinely saved or analyzed because of the human hours required to process them. So to begin applying SPR to X-ray-guided procedures, the researchers first had to create their own training dataset, harnessing the power of synthetic data and deep neural networks to simulate surgical workflows and X-ray sequences based on a preexisting database of annotated CT scan images. They simulated enough data to successfully train their own machine learning-powered SPR algorithm specifically for X-ray sequences.
“We simulated not only the visual appearance of images but also the dynamics of surgical workflows in X-ray to provide a viable alternative to real image sources—and then we set out to show that this approach transfers to the real world,” Killeen said.
The researchers validated their novel approach in cadaver experiments and successfully demonstrated that the Pelphix workflow can be applied to real-world X-ray-based SPR algorithms. They suggest that future algorithms use Pelphix’s simulations for pretraining before fine-tuning on real image sequences from actual human patients.
The team is now collecting patient data for a large-scale validation effort.
“The next step in this research is to refine the workflow structure based on our initial results and deploy more advanced algorithms on large-scale datasets of X-ray images collected from patient procedures,” Killeen said. “In the long term, this work is a first step toward obtaining insights into the science of orthopedic surgery from a big data perspective.”
The researchers hope that Pelphix’s success will motivate the routine collection and interpretation of X-ray data to enable further advances in surgical data science, ultimately improving the standard of care for patients.
“In some ways, modern operating theaters and the surgeries happening within them are much like the expanding universe, in that 95% of it is dark or unobservable,” says senior co-author Mathias Unberath, an assistant professor of computer science affiliated with the Malone Center, the principal investigator of the ARCADE Lab, and Killeen’s advisor. “That is, many complex processes happen during surgeries: tissue is manipulated, instruments are placed, and sometimes, errors are made and—hopefully—corrected swiftly. However, none of these processes is documented precisely. Surgical data science and surgical phase recognition—and approaches like Pelphix—are working to make that inscrutable 95% of surgery data observable, to patients’ benefit.”
Additional co-authors include Russell Taylor, John C. Malone Professor of Computer Science and the director of the Laboratory for Computational Sensing and Robotics; Greg Osgood, an associate professor of orthopaedic surgery and chief of orthopaedic trauma in the Johns Hopkins Department of Orthopaedic Surgery; Mehran Armand, a professor of orthopaedic surgery with joint appointments in the departments of mechanical engineering and computer science; Jan Mangulabnan, a doctoral candidate in the Department of Computer Science and a member of the ARCADE Lab; and biomedical engineering graduate student Han Zhang.