Accessing the brain for neurosurgery involves drilling and cutting that can cause deep-brain anatomy to shift or become distorted. This can create discrepancies between pre-operative imaging and the actual state of the brain during a procedure.
Current surgical navigation systems may assist in providing live guidance, but typically use pins and clamps to hold a patient’s head firmly in place, which carries the risk of complications and can prolong recovery time.
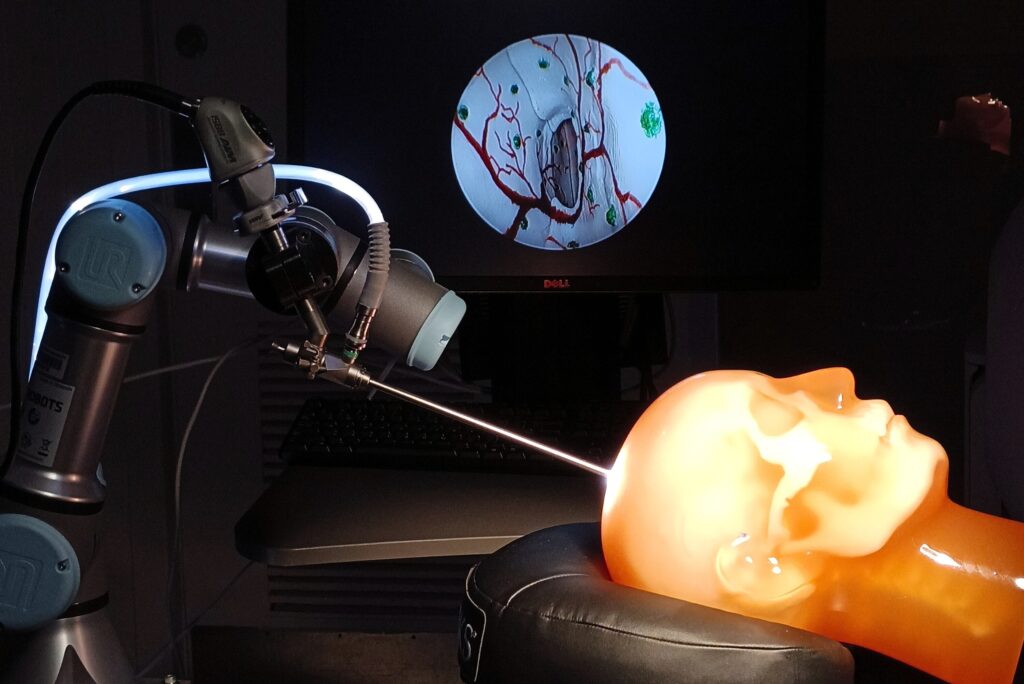
A team of researchers at the Imaging for Surgery, Therapy, and Radiology (I-STAR) Labs at Johns Hopkins University is working on a less invasive solution that doesn’t require additional equipment or expose patients to the extra radiation or long scan times typically associated with live imaging. They have partnered with medical device company Medtronic and the National Institutes of Health to develop a real-time guidance system that uses an endoscope, a piece of equipment already commonly employed in neurosurgeries.
Their work is described in in IEEE Transactions on Medical Robotics and Bionics.
“Our study demonstrates the superiority of this real-time 3D navigation method over current visualization techniques,” says Prasad Vagdargi, a doctoral candidate at the I-STAR Labs and primary author of the study. Vagdargi is advised by Jeffrey H. Siewerdsen, John C. Malone Professor of Biomedical Engineering, and Gregory D. Hager, Mandell Bellmore Professor of Computer Science.
The team’s surgical guidance method builds on an advanced computer vision technique called simultaneous localization and mapping, or SLAM, which has also been used for navigation in self-driving cars. After calibrating an endoscopic video feed, the team’s SLAM algorithm tracks important visual details in each frame and uses those details to determine where the endoscope camera is and how it’s positioned. The algorithm then transforms those details into a 3D model of the object—in this case, the inside of a patient’s skull. This model is then overlaid with the real-world video feed to visualize targeted structures on-screen in real time.
“Think of it as a dynamic 3D map of a patient’s brain that you can use to track and match deep brain deformation with preoperative imaging,” Vagdargi says. “Combining this map with an augmented reality overlay on the endoscopic video will help surgeons to visualize targets and critical anatomy well beyond the surface of the brain.”
The researchers have coined the term “augmented endoscopy” for this highly accurate, state-of-the-art method of live surgical visualization. Through a series of preclinical experiments, they demonstrated that this method is more than 16 times faster than previous computer vision techniques while still maintaining submillimeter accuracy.
The team predicts that the improved accuracy afforded by augmented endoscopy may lead to reduced complications, shorter operation times, and therefore increased surgical efficiency—not only in neurosurgeries but also in other endoscopic procedures across medical disciplines.
The research team is currently collaborating with neurosurgeons at the Johns Hopkins Hospital in a clinical study with the goal of refining and validating their method for use in real operating rooms. The researchers are also expanding their approach by applying artificial intelligence and machine learning methods to further improve the speed and precision of future iterations of their algorithm.
“We believe our findings are very important and feel that augmented endoscopy is a technology whose time has now come,” says Vagdargi.
Study co-authors include Craig Jones, an assistant research professor in the Department of Computer Science; colleagues in the Whiting School’s Department of Biomedical Engineering and the Department of Neurology and Neurosurgery at the Johns Hopkins Hospital; and industry collaborators at Medtronic.
This article originally appeared on the Hub >>
Image Caption: A robot arm with a neuroendoscope positioned in relation to a human head “phantom” for the experimental validation of an augmented neuroendoscopy system.